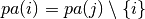DAG
Copying
|
Return a copy of the current DAG. |
|
Rename the nodes in this graph according to |
|
Return the induced subgraph over only |
Information about nodes
|
Return all nodes that are parents of the node or set of nodes |
|
Return all nodes that are children of the node or set of nodes |
|
Return all nodes that are adjacent to the node or set of nodes |
|
Return the Markov blanket of |
|
Return the ancestors of |
|
Return the descendants of |
|
Return the indegree of |
|
Return the outdegree of |
|
Return all arcs with target |
|
Return all arcs with source |
|
Return all arcs with |
Graph modification
|
Add |
|
Add nodes to the graph from the collection |
|
Remove the node |
|
Add the arc |
|
Add arcs to the graph from the collection |
|
Remove the arc |
|
Reverse the arc |
Graph properties
|
Check if this DAG has an arc |
Get all nodes in the graph that have no parents. |
|
Get all nodes in the graph that have no children. |
|
Get all reversible (aka covered) arcs in the DAG. |
|
|
Check if the arc |
Get all arcs in the graph that participate in a v-structure. |
|
Get all v-structures in the graph, i.e., triples of the form (i, k, j) such that |
|
Return all triples of the form ( |
|
Return the set of nodes which in |
Ordering
Return a topological sort of the nodes in the graph. |
|
|
Check that |
|
Return the number of "errors" in |
Comparison to other DAGs
|
Compute the structural Hamming distance between this DAG and the DAG |
|
Compute the structure Hamming distance between the skeleton of this DAG and the skeleton of the graph |
|
Check if this DAG is (interventionally) Markov equivalent to the DAG |
|
Check if this DAG is an IMAP of the DAG |
|
Check if this DAG is a minimal IMAP of other, i.e., it is an IMAP and no proper subgraph of this DAG is an IMAP of other. |
|
Return the total number of edge reversals plus twice the number of edge additions/deletions required to turn this DAG into the DAG |
|
Return the "confusion matrix" associated with estimating the CPDAG of |
Return the "confusion matrix" associated with estimating the skeleton of |
Separation statements
|
Check if |
|
Find all nodes d-separated from |
|
Check if the distribution of |
Return the local Markov statements of this DAG, i.e., those of the form |
Conversion to/from other formats
|
Return a DAG with arcs given by |
|
Return an adjacency matrix for this DAG. |
|
Convert a networkx DiGraph into a DAG. |
Convert DAG to a networkx DiGraph. |
|
Create a DAG from a dataframe, where the indices and columns are node names and a nonzero entry indicates the presence of an edge. |
|
|
Turn this DAG into a dataframe, where the indices and columns are node names and a nonzero entry indicates the presence of an edge. |
Conversion to other graphs
Return the (undirected) moral graph of this DAG, i.e., the graph with the parents of all nodes made adjacent. |
|
|
Return the maximal ancestral graph (MAG) that results from marginalizing out |
Return the completed partially directed acyclic graph (CPDAG, aka essential graph) that represents the Markov equivalence class of this DAG. |
|
|
Return the interventional essential graph (aka CPDAG) associated with this DAG. |
Chickering Sequences
|
Return the nodes in this graph which are "resolved sinks" with respect to the graph |
|
Return a Chickering sequence from this DAG to an I-MAP |
|
Identify an edge operation (covered edge reversal or edge addition) which decreases the Chickering distance from this DAG to |
Directed Clique Trees
|
Return the directed clique tree associated with this DAG. |
Return the contracted directed clique tree associated with this DAG. |
|
Return the residuals associated with this DAG. |
|
Return the residual essential graph associated with this DAG. |
Intervention Design
|
Find the smallest set of interventions which fully orients the CPDAG into this DAG. |
Greedily pick |
|
Find a set of interventions which fully orients a CPDAG into this DAG, using greedy selection of the interventions. |